Methodology
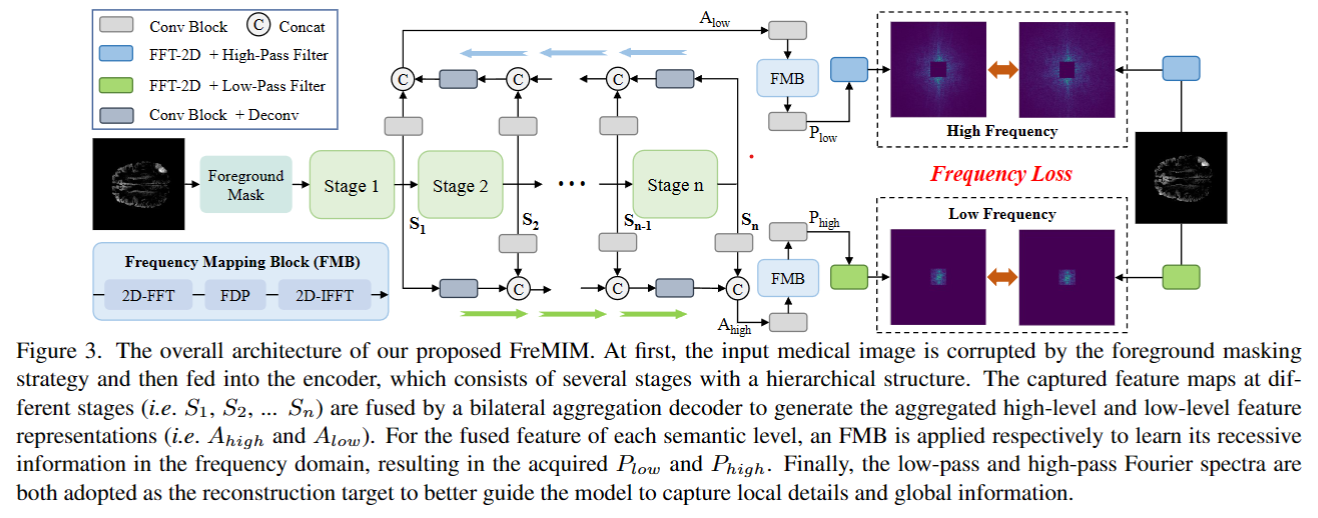
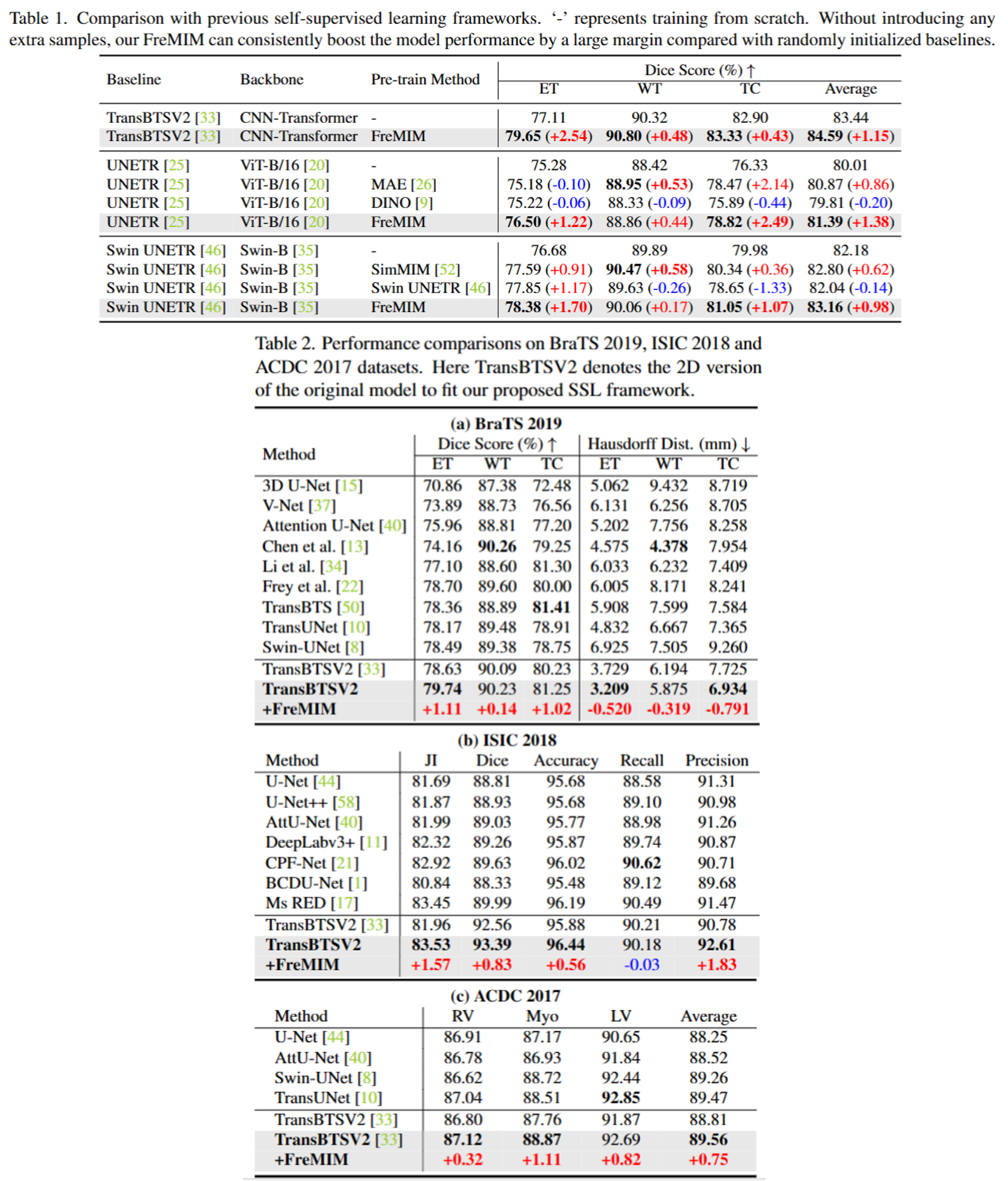
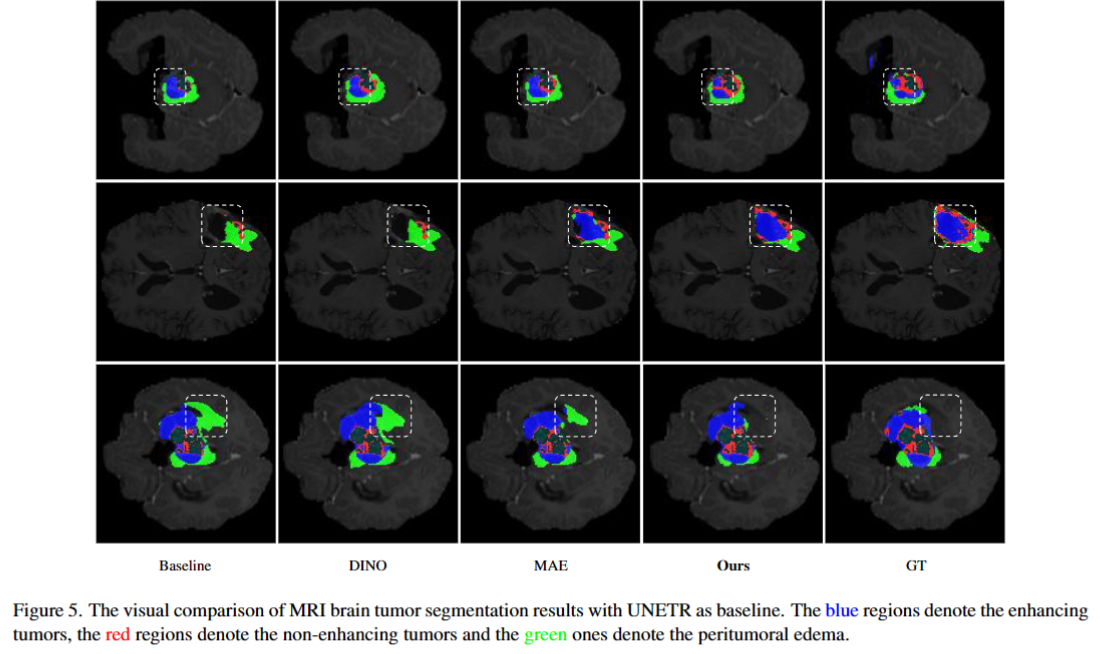
The research community has witnessed the powerful potential of self-supervised Masked Image Modeling (MIM), which enables the models capable of learning visual representation from unlabeled data. In this paper, to incorporate both the crucial global structural information and local details for dense prediction tasks, we alter the perspective to the frequency domain and present a new MIM-based framework named FreMIM for self-supervised pre-training to better accomplish medical image segmentation tasks. Based on the observations that the detailed structural information mainly lies in the high-frequency components and the high-level semantics are abundant in the low-frequency counterparts, we further incorporate multi-stage supervision to guide the representation learning during the pre-training phase. Extensive experiments on three benchmark datasets show the superior advantage of our FreMIM over previous state-of-the-art MIM methods. Compared with various baselines trained from scratch, our FreMIM could consistently bring considerable improvements to model performance.

@inproceedings{wang2024fremim,
title={FreMIM: Fourier Transform Meets Masked Image Modeling for Medical Image Segmentation},
author={Wang, Wenxuan and Wang, Jing and Chen, Chen and Jiao, Jianbo and Cai, Yuanxiu and Song, Shanshan and Li, Jiangyun},
booktitle={Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision},
pages={7860--7870},
year={2024}
}